
Participate in the ESHI-MICCAI machine learning challenge!
ESHI, in collaboration with MICCAI, is proud to announce the first autoPET challenge on automated tumor lesion segmentation on whole-body FDG-PET/CT. The aim of the challenge is to foster and promote research on machine learning-based automation and data evaluation.
AutoPET provides a large-scale, publicly available dataset of 1014 studies of 900 patients acquired on a single site. The task is to develop a machine-learning algorithm for accurate lesion segmentation of whole-body FDG-PET/CT while avoiding false positives (brain, bladder, etc.).
The training data is licensed as CC BY, i.e. everyone (also non-participants of the challenge) are free to use the training data set in their respective work, given attribution in the publication.
The 7 highest ranking submissions will compete for a total of 15’000€ and will be invited to present their methods at MICCAI 2021. Subsequently, they are invited to write a peer-reviewed journal paper that performs a full analysis of the results and highlights the key findings and methods.
All details about the challenge can be found below and via the official challenge page.

Figure: Example case of fused FDG-PET/CT whole-body data. The right image shows the manually segmented malignant lesions.
Introduction
Positron Emission Tomography / Computed Tomography (PET/CT) is an integral part of the diagnostic workup for various malignant solid tumor entities. Due to its wide applicability, Fluorodeoxyglucose (FDG) is the most widely used PET tracer in an oncological setting reflecting glucose consumption of tissues, e.g. typically increased glucose consumption of tumor lesions.
As part of the clinical routine analysis, PET/CT is mostly analyzed in a qualitative way by experienced medical imaging experts. Additional quantitative evaluation of PET information would potentially allow for more precise and individualized diagnostic decisions.
A crucial initial processing step for quantitative PET/CT analysis is segmentation of tumor lesions enabling accurate feature extraction, tumor characterization, oncologic staging and image-based therapy response assessment. Manual lesion segmentation is however associated with enormous effort and cost and is thus infeasible in clinical routine. Automation of this task is thus necessary for widespread clinical implementation of comprehensive PET image analysis.
Recent progress in automated PET/CT lesion segmentation using deep learning methods has demonstrated the principle feasibility of this task. However, despite these recent advances tumor lesion detection and segmentation in whole-body PET/CT is still a challenging task. The specific difficulty of lesion segmentation in FDG-PET lies in the fact that not only tumor lesions but also healthy organs (e.g. the brain) can have significant FDG uptake; avoiding false positive segmentations can thus be difficult. One bottleneck for progress in automated PET lesion segmentation is the limited availability of training data that would allow for algorithm development and optimization.
To promote research on machine learning-based automated tumor lesion segmentation on whole-body FDG-PET/CT data we host the autoPET challenge and provide a large, publicly available training data set.
AutoPET is hosted at the MICCAI 2022:
Dataset
The challenge cohort consists of patients with histologically proven malignant melanoma, lymphoma or lung cancer as well as negative control patients who were examined by FDG-PET/CT in two large medical centers (University Hospital Tübingen, Germany & University Hospital of the LMU in Munich, Germany).
All PET/CT data within this challenge have been acquired on state-of-the-art PET/CT scanners (Siemens Biograph mCT, mCT Flow and Biograph 64, GE Discovery 690) using standardized protocols following international guidelines. CT as well as PET data are provided as 3D volumes consisting of stacks of axial slices. Data provided as part of this challenge consists of whole-body examinations. Usually, the scan range of these examinations extends from the skull base to the mid-thigh level. If clinically relevant, scans can be extended to cover the entire body including the entire head and legs/feet.
You can find more details about the PET/CT acquisition protocol, the training and test cohort and the annotation on the official challenge page:
Task
Automatic tumor lesion segmentation in whole-body FDG-PET/CT on large-scale database of 1014 studies of 900 patients (training database) acquired on a single site:
- accurate and fast lesion segmentation
- avoidance of false positives (brain, bladder, etc.)
Testing will be performed on 200 studies (held-out test database) with 100 studies originating from the same hospital as the training database and 100 are drawn from a different hospital with similar acquisition protocol to assess algorithm robustness and generalizability.
Rules
To participate, you have to agree to the following rules:
- Submission is in the form of docker containers, as we can’t allow access to the test images during the competition. You will be given a template docker container to use for this.
- Participants are requested to publish a (brief) description of their method and results on ArXiv (or similar pre-print platforms) that is linked to their final submission. There is no page limit for that description, but it has to include a conclusive description of the approach of the participating team.
- Training data is licensed as CC BY, i.e. everyone (also non-participants of the challenge) are free to use the training data set in their respective work, given attribution in the publication.
- Algorithms must be made publicly available as e.g. GitHub repository with a permissive open source license (Apache Licence 2.0, MIT Licence, GNU GPLv3, GNU AGPLv3, Mozilla Public Licence 2.0, Boost Software Licence 1.0, The Unlicence)
- Researchers belonging to the institutes of the organizers are not allowed to participate to avoid potential conflict of interest.
Publication policy
-
Participants may publish papers including their official performance on the challenge data set, given proper reference of the challenge and the dataset. There is no embargo time in that regard. Please use the following references:Database: DOI_WILL_FOLLOW_SHORTLY
- We aim to publish a summary of the challenge in a peer-reviewed journal (further information). The first and last author of the submitted arxiv paper will qualify as authors in the summary paper. Participating teams are free to publish their own results in a separate publication after coordination (for selected submissions of summary paper) to avoid siginficant overlap with the summary paper.
🕑 1st April 2022
Challenge opens, release of training data
🕑 25th April 2022
Submission system opens
🕑 31st August 2022, 23:59 pm (UTC+1)
Challenge closes
🕑 18th – 22nd September 2022
MICCAI
Top 7 performing submissions will present their methods
🕑 Beginning of October
Top 7 performing teams will be invited to contribute to draft and submit a manuscript describing the methods and results of the challenge in a peer-reviewed journal
Evaluation criteria
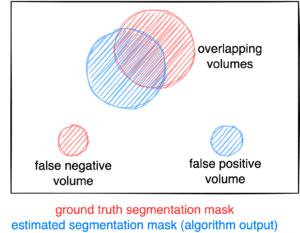
Figure: Example for the evaluation. The Dice score is calculated to measure the correct overlap between predicted lesion segmentation (blue) and ground truth (red). Additionally special emphasis is put on false positives by measuring their volume, i.e. large false positives like brain or bladder will result in a low score and false negatives by measuring their volume (i.e. entirely missed lesions).
Evaluation will be performed on held-out test cases of 200 patients. Test cases are split in two subgroups: 100 are drawn from the same hospital as the training cases (University Hospital Tübingen, Germany) and 100 are drawn from a different hospital (University Hospital of LMU in Munich, Germany) with similar acquisition protocols.
A combination of two metrics reflecting the aims and specific challenges for the task of PET lesion segmentation:
- Foreground Dice score of segmented lesions
- Volume of false positive connected components that do not overlap with positives (=false positive volume)
- Volume of positive connected components in the ground truth that do not overlap with the estimated segmentation mask (=false negative volume)
In case of test data that do not contain positives (no FDG-avid lesions), only metric 2 will be used.
A python script computing these evaluation metrics is provided under https://github.com/lab-midas/autoPET.
Challenge Publication
Participants must submit a method description along with their submission on the final test set. This should be in Springer Lecture Notes in Computer Science format [link to Overleaf template]. The paper should highlight the main steps taken to arrive at the method used on the final test set. In particular, the following should be described:
- Data preprocessing and augmentation
- Method description
- Post Processing
- Results
- Link to public code repository
Each method description paper should be uploaded to a pre-print platform such as ArXiv, so that the paper can be easily linked to the final leaderboard. Each final submission needs to be linked to a pre-print paper. More information on this can be found in the submission instructions.
Post Challenge Publication
After the challenge has finished, we will invitate the Top 7 performing submissions to write a peer-reviewed journal paper that performs a full analysis of the results and highlights the key findings and methods. Further method submissions may be invited to the summary paper. The first and last author of the submitted arxiv paper will qualify as authors in the summary paper. The participating teams may publish their own results separately after coordination (for selected submissions of summary paper) to avoid siginficant overlap with the summary paper. More information on this will be provided on conclusion of the challenge.
Ranking
The submitted algorithms will be ranked according to:
Step 1: Separate rankings will be computed based on each metric (for metric 1: higher Dice score = better, for metrics 2 and 3: lower volumes = better)
Step 2: From the three ranking tables, the mean ranking of each participant will be computed as the numerical mean of the single rankings (metric 1: 50 % weight, metrics 2 and 3: 25 % weight each)
Step 3: In case of equal ranking, the achieved Dice metric will be used as a tie break.
Prizes
- 1st place: 6,000 € 🥇
- 2nd place: 3,000 € 🥈
- 3rd place: 2,000 € 🥉
- 4rd-7th place: 1,000 € (each) 🏆
This challenge is hosted at the Medical Image Computing and Computer Assisted Intervention (MICCAI) 2022 and supported by the European Society for hybrid, molecular and translational imaging (ESHI)


Medical Image and Data Analysis (MIDAS.lab)



